BLAT
IGV supports BLAT (BLAST-like Alignment Tool) for on-the-fly alignment of query sequences up to 8 kb in length.
How to run BLAT#
There are several options for running a BLAT query depending on the source of the query sequence:
- User-specified sequence
- Select Tools > BLAT and enter the nucleotide sequence into the window that pops up.
- Aligned read sequence
- Right-click on the aligned read and select BLAT read sequence from the popup menu.
- If soft clips are displayed and are of sufficient length, the popup menu will also include options to BLAT the soft-clipped sequence.
- Reference sequence defined by a feature
- Right-click on a feature in a feature track and select BLAT sequence from the popup menu. The BLAT input sequence is the section of the reference genome defined by the feature start and end bounds.
- Reference sequence defined by a region of interest (ROI)
- After creating a region of interest, click on the region's red bar and select BLAT sequence from the popup menu. The BLAT input sequence is the section of the reference genome defined by the region bounds.
The sequence query is sent to an external BLAT search engine. The default search engine is the BLAT server hosted at the UCSC Genome Browser. UCSC's BLAT search supports most UCSC derived genomes including human and mouse genomes. See below for instructions on specifying a custom BLAT server, or configuring a command-line BLAT tool.
BLAT results#
The results from a BLAT query are presented in a new feature track that is added to the lower panel of the IGV window and a results panel that is displayed in a separate popup window.
Feature track#
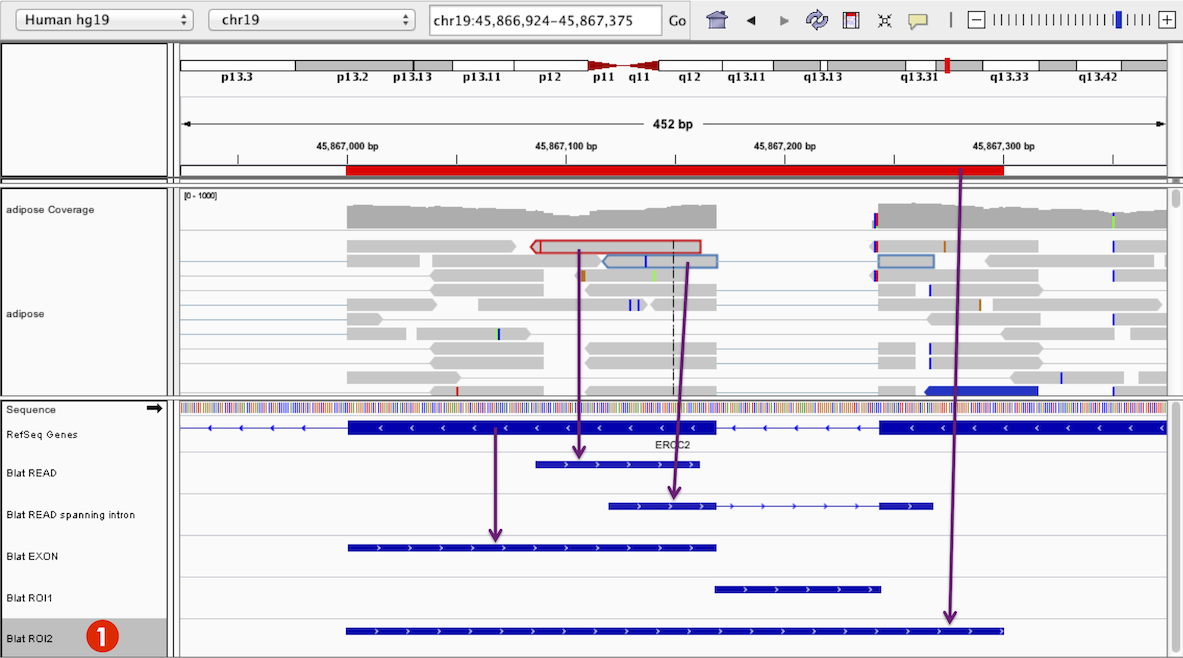
The results of each BLAT search appear as a new feature track in the lower panel of IGV's display, where each feature in the track represents a hit. The screenshot below shows five different BLAT feature tracks resulting from queries for the following sequences:
- The red highlighted read
- The blue highlighted RNA-seq read spanning an intron
- An exon feature
- A region of interest (ROI) covering an intronic region
- A region of interest (ROI) spanning the region that covers the other four examples.
The resulting hits are displayed as blue rectangles and lines are used to represent any gaps in the alignment, as can be seen in the results of the BLAT of the RNA-seq read sequence. The alignment score is represented as opacity resulting in a dark blue to light blue color. The arrowheads represent the directionality relative to the original search sequence.
By default, result tracks for BLAT searches of a read sequence in an alignment track will be assigned a track name that is the same as the read name. Result tracks for other BLAT searches will be named simply "BLAT". In the example below, the user has renamed all the BLAT output tracks. This is done by right-clicking on a track and selecting Rename Track from the popup menu. For example, the track marked 1 is now named Blat ROI2.
BLAT result tracks can be manipulated just like any other feature track via the popup menu.
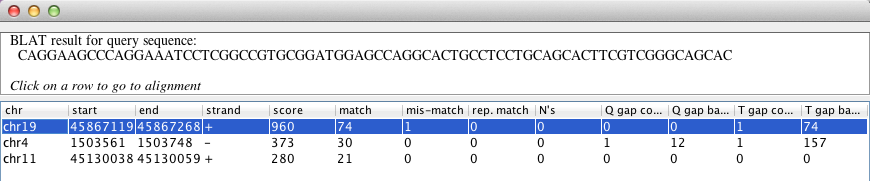
Results panel#
Each BLAT search presents a separate results panel displayed in a popup window. The query sequence is displayed at the top, and a then row for every hit, including the location of the hit, match score, and other metrics as shown in the screenshot below. Hits are listed in descending order of the score.
If you close the results window, you can reopen it by right-clicking on the corresponding feature track in the IGV window and selecting Open table view from the popup menu.
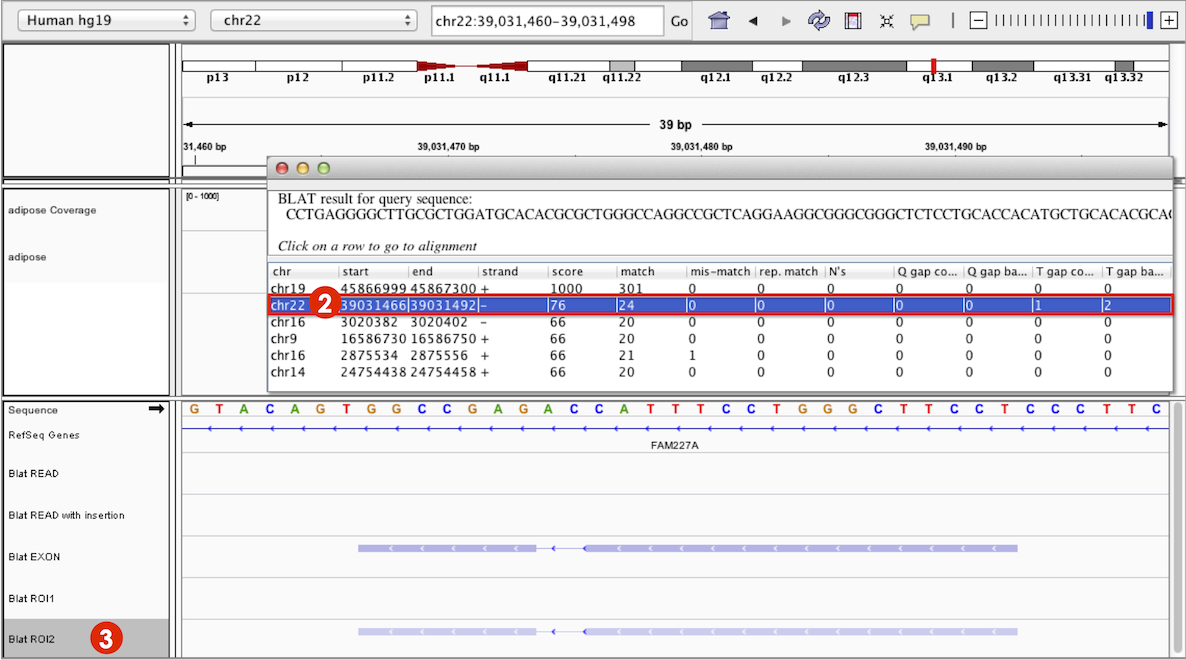
Click on a row in the results panel to navigate the IGV view to the selected result locus. In the example screenshot below, clicking on the second row (marked 2) in the table, has changed the IGV view to the hit locus on chromosome 22. In the corresponding feature track in the main IGV window (Blat ROI2 marked 3), you can see the feature is a lighter blue than the feature on chromosome 19 (see the same track Blat ROI2 marked 1 in the screenshot above). This is because the locus of the original query sequence on chromosome 19 has a higher alignment score than the hit found on chromosome 22.
Customizing BLAT#
By default, IGV uses a public BLAT web service hosted at UCSC, and BLAT support is limited to reference genomes that are available at UCSC. However, the BLAT utility used by IGV can be changed to another BLAT web service or a locally executable command line program that returns BLAT-like results.
To change to a different BLAT utility, select
View > Preferences, click on the Advanced tab, and edit the BLAT URL field. The value is a string template with placeholders for the
search sequence $SEQUENCE and genome ID $DB. These values are substituted at runtime. The default
value is
https://genome.ucsc.edu/cgi-bin/hgBlat?userSeq=$SEQUENCE&type=DNA&db=$DB&output=json
Output from the web service or command line program should be JSON containing a blat property at the top level,
followed by an array of PSL records representing the alignments. Each PSL record is represented as a JSON array. An example is given below.
Details of the PSL format is available at UCSC.
{
"blat" : [
[33,1,0,0,0,0,0,0,"+","YourSeq",40,0,34,"chr1",249250621,155161117,155161151,1, 34,0,155161117],
[33,1,0,0,0,0,0,0,"+","YourSeq",40,0,34,"chr1",249250621,155161255,155161289,1, 34,0,155161255],
[33,1,0,0,0,0,0,0,"+","YourSeq",40,0,34,"chr1",249250621,155161315,155161349,1, 34,0,155161315],
[33,1,0,0,0,0,0,0,"+","YourSeq",40,0,34,"chr1",249250621,155161435,155161469,1, 34,0,155161435],
[33,1,0,0,0,0,0,0,"+","YourSeq",40,0,34,"chr1",249250621,155161495,155161529,1, 34,0,155161495]
]
}
If configuring a local command line program, the output should be written to STDOUT. A simple example of configuring a command line BLAT shell script, written for Linux and Mac systems, is given below. This script takes the search sequence and database ID as arguments, and simply calls the UCSC service referenced above and diverts the output to STDOUT.
The script testBlat.sh:
#!/bin/bash
wget -q -O - --no-check-certificate "https://genome.ucsc.edu/cgi-bin/hgBlat?userSeq=$1&type=DNA&db=$2&output=json"
To use the script, set the BLAT URL IGV preference to:
testBlat.sh $SEQUENCE $DB