Motif finder
The motif finder searches for a specified nucleotide sequence in the reference genome. The results are displayed as features in two new tracks.
How to run the motif finder#
-
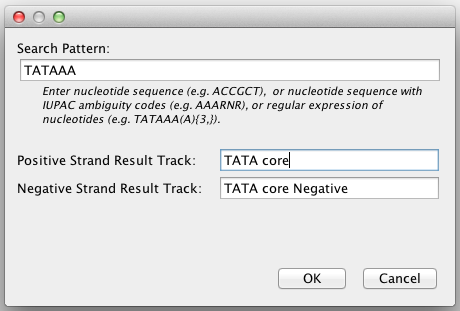
Select Tools > Find Motif to bring up the motif finder dialog.
-
Enter the sequence for which to search, using one of following three formats:
-
A sequence of nucleotides.
-
A sequence of nucleotides with IUPAC ambiguity codes.
- For example, let's say you want to find bacterial promoter upstream elements consisting of 6 adenines (A), followed by a purine (A or G), then any nucleotide (A, C, G, or T), and finally another purine (A or G). You would enter the sequence "AAAAAARNR".
-
A regular expression that follows Java regex syntax.
- For example, to find occurrences of the canonical TATA box sequences TATAAAAA, TATATAAA, and TATAAATA, you can enter the regular expression "TATA[AT]A[AT]A". Regular expressions are particularly useful for finding variable length sequences. For example, to search for the sequence TATAAA, optionally followed by any number of additional adenines, enter the regular expression "TATAAA+".
-
-
Enter names for the result feature tracks that will show where the sequence matches the positive and negative strands of the reference genome.
- By default, the results from the positive strand are displayed in blue, and results from the negative strand in red. To change the color, right-click on the feature track and select Change Track Color from the pop-up menu.
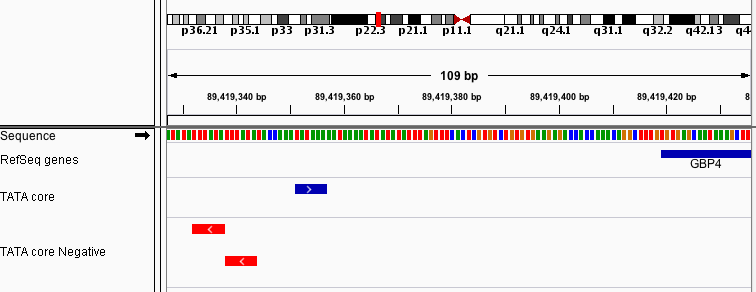
Example#
Since we entered a short sequence, it gets a large number of hits. Looking at the results directly upstream of the gene GBP4, we see a match on the positive strand and two on the negative strand.
By default, the search result tracks are displayed in Expanded mode, so you can see overlapping matches.