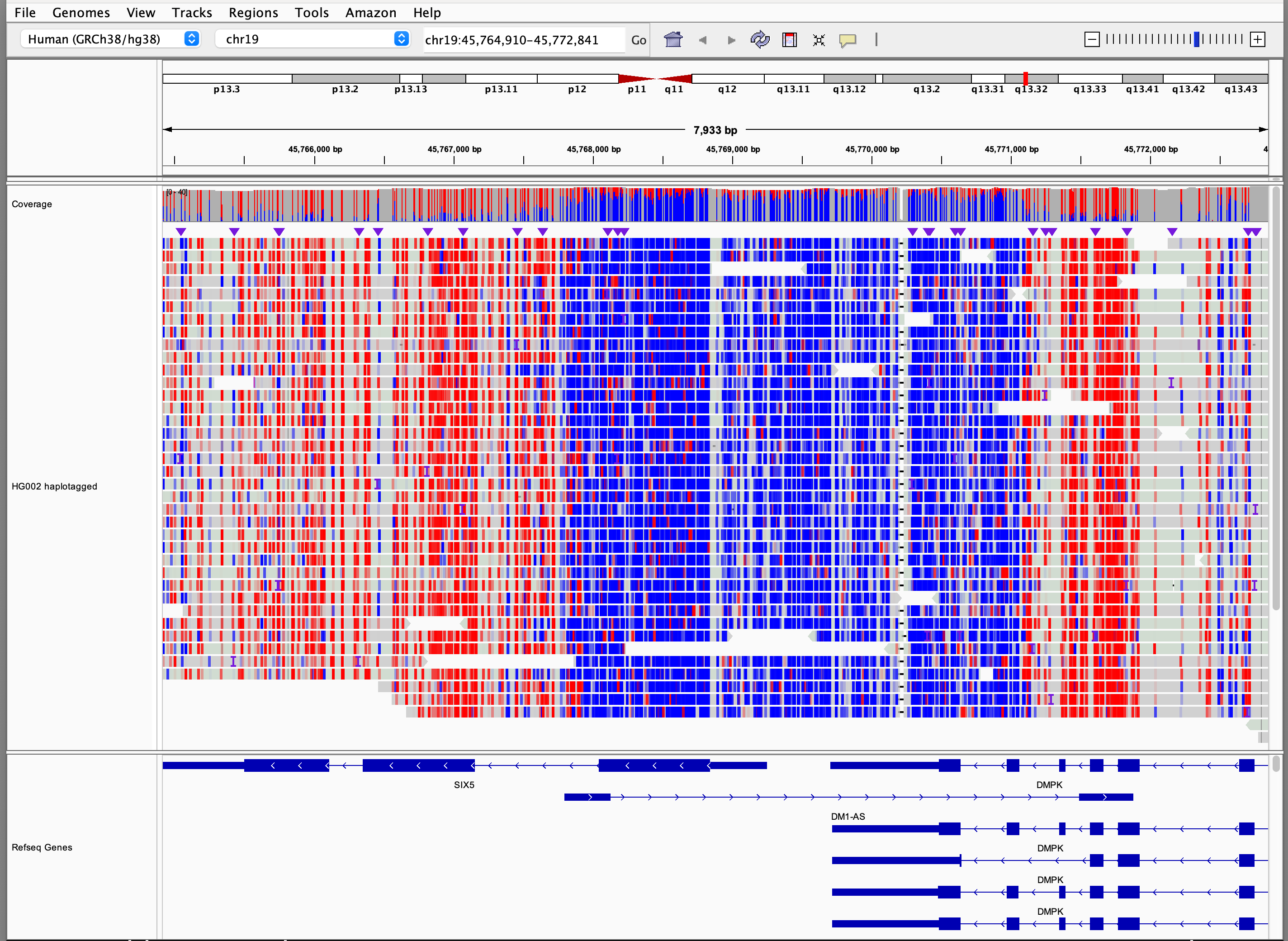
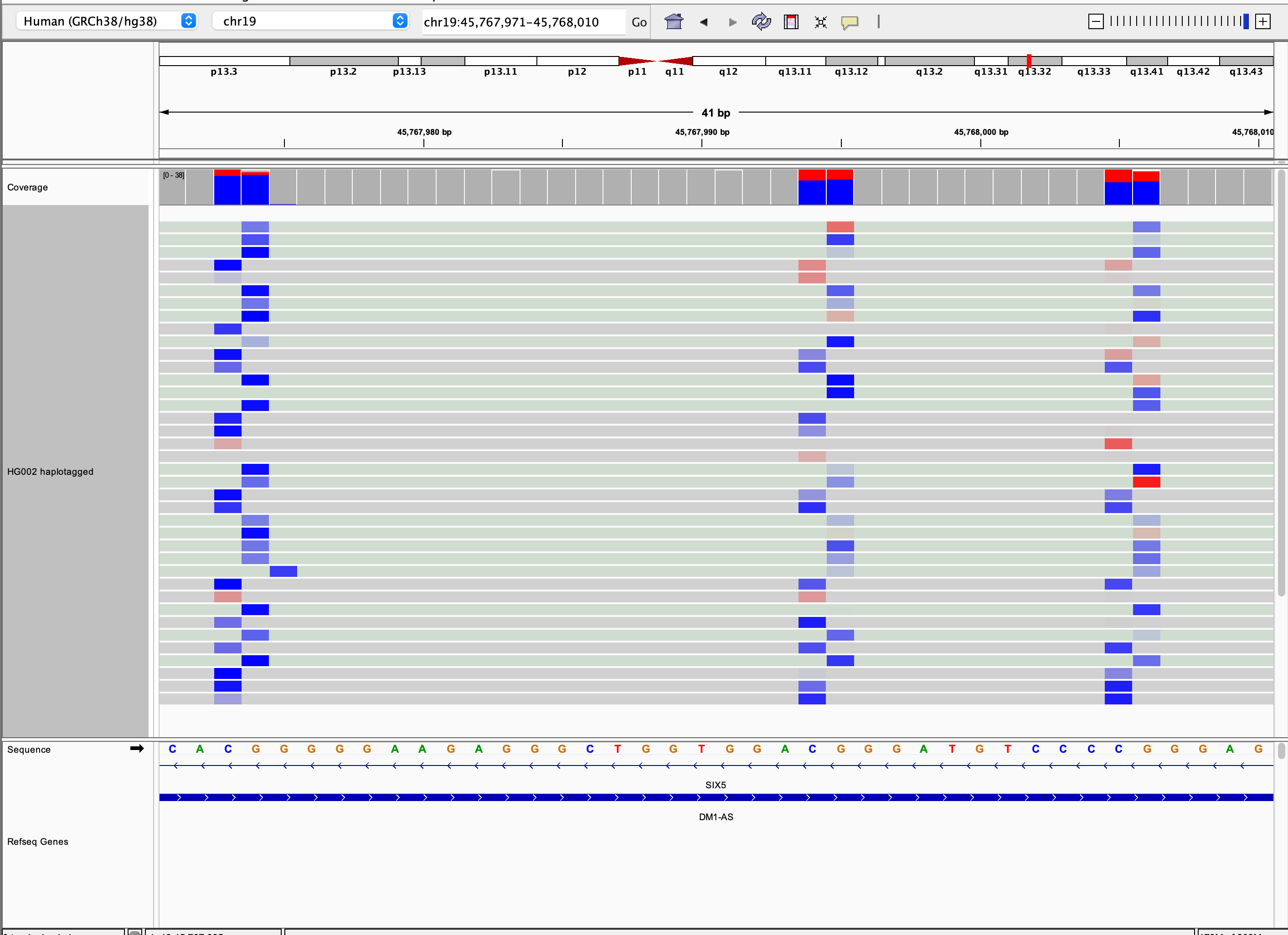
Base modifications
Options for coloring alignments by base modifications are enabled for alignments with MM and optionally ML tags,
denoting modification type and likelihood respectively (see SAM/BAM format tag specifications).
When these tags are present options appear in the Color by section of the alignment track menu to color
all modifications present, or a specific modification, using either a mono-color (default) or 2-color scheme. For example,
in an experiment with both 5mC and 5hmC modifications the following menu options are available.

Display conventions#
Display conventions for base modification coloring options are listed below.
-
Alignment blocks are colored by DNA strand. For 3rd generation reads this is the read strand. For paired reads, the DNA strand is assumed to be the strand of the read marked first-in-pair.
- Grey for forward read
- Sage for reverse read
-
Alignments are grouped by DNA strand. This can be disabled from the Base Mods tab of the preferences window.
-
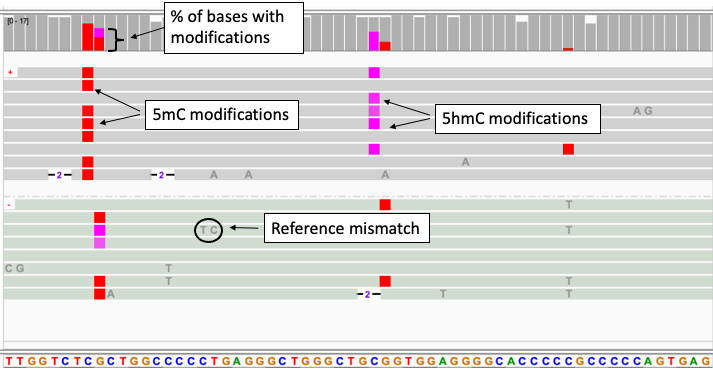
Mismatches to reference are flagged by displaying the read base in dark grey.
-
Base modifications are indicated by color, with a distinct color assigned to each modification type. These colors are settable in user preferences
-
Alpha transparency is used to indicate the likelihood of the modification as reported by the ML tag.
In both modes a base is assigned a color if the modification likelhiood exceeds a user-settable threshold, 0.5 (50%),
by default. In the case of multiple modifications of a single base, the modification with the highest likelihood is
used to assign color if that likelihood exceeds the threshold, If no modification likelihood exceeds the threshold
the base remains uncolored.
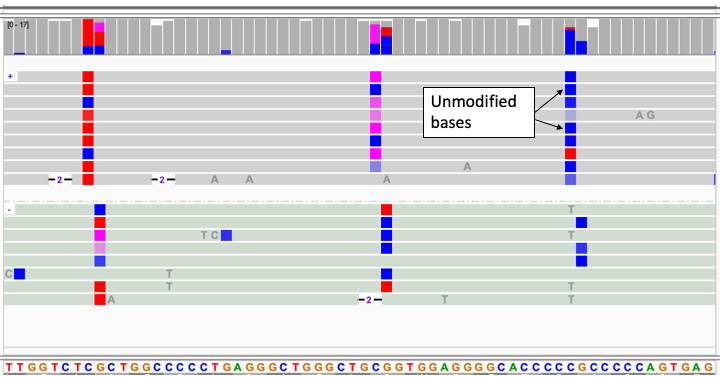
The two-color option extends the mono-color scheme by assigning a color for the unmodified base, blue by default. The likelihood of no modification is taken as 1 - (sum of all modification likelhoods). For example, a base with likelihoods of 5hmC = 30% and 5mC = 10%, would be assigned a likelihood of no modification of 60%. This mode gives equal emphasis to modified and unmodified bases.
Note that visualization of 5-Methylcytosine (5mC) modifications in 2-color mode, which assigns red to 5mC modification
and blue to a likely unmodified base, is consistent
with visualization of methylation from bisulfite sequencing, making experiments from the
two technologies comparable.
Default colors for common modifications are show below. The colors are user settable on the Base Mods tab of the user
preferences window. Additionally, color representing the unmodified base can be set and
customized for each canonical base (A,C,T, and G).

Example screenshots#
- Mono-color display of reads with 5mC and 5hmC modifications
- Two-color display of reads with 5mC and 5hmC modifications
- 5mC methylation
5mC experiment in 2-Color mode (5mC == Red, No modification == Blue, Threshold = 0.5)